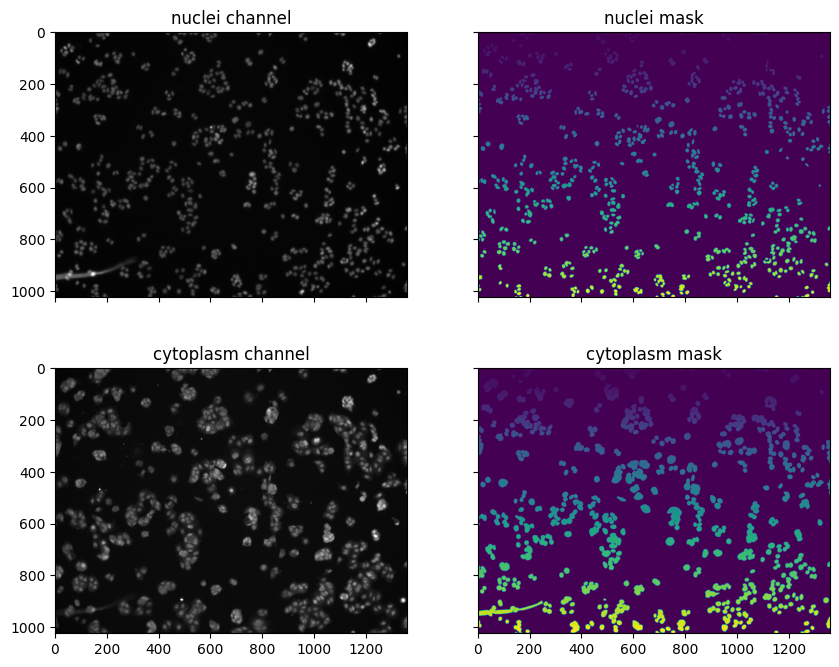
cellpose-segmentation pipeline) results in masks that identify where cell bodies and/or nuclei are located in an image. Once you have obtained masks, you can use them to quantify the concentration of the labeled molecules within the segmented regions using tools such as the scikit-image package in Python.
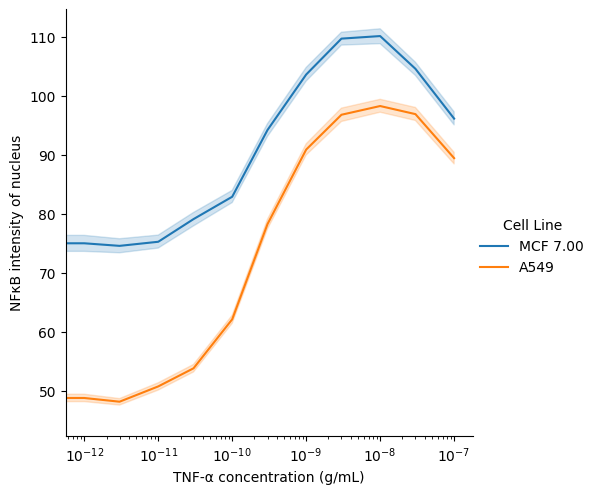
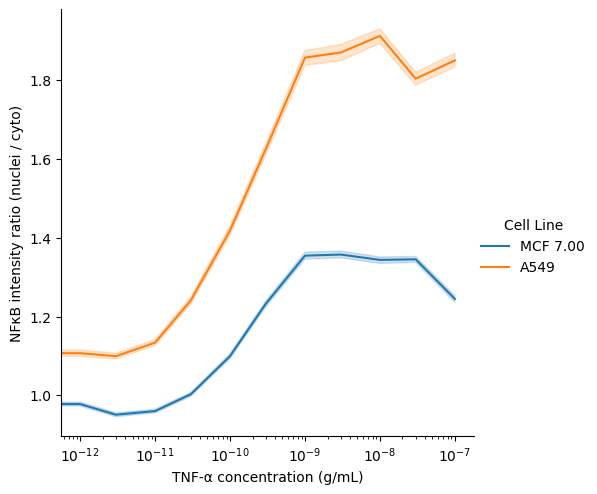
In this notebook, we compare the quantity of a protein of interest (transcription factor NFκB) in the cytoplasm versus the nucleus of cell in response to varying doses of TNF-α. The microscopy data were originally from the BBBC014v1 dataset, available from the Broad Bioimage Benchmark Collection.
Ljosa, V., Sokolnicki, K. L., & Carpenter, A. E. (2012). Annotated high-throughput microscopy image sets for validation. Nature Methods, 9(7), 637-637.Using scikit-image, we read in the images and masks and extract the mean intensity of each labeled region. Using Pandas, we filter for segmented cells that contain exactly one segmented nucleus.