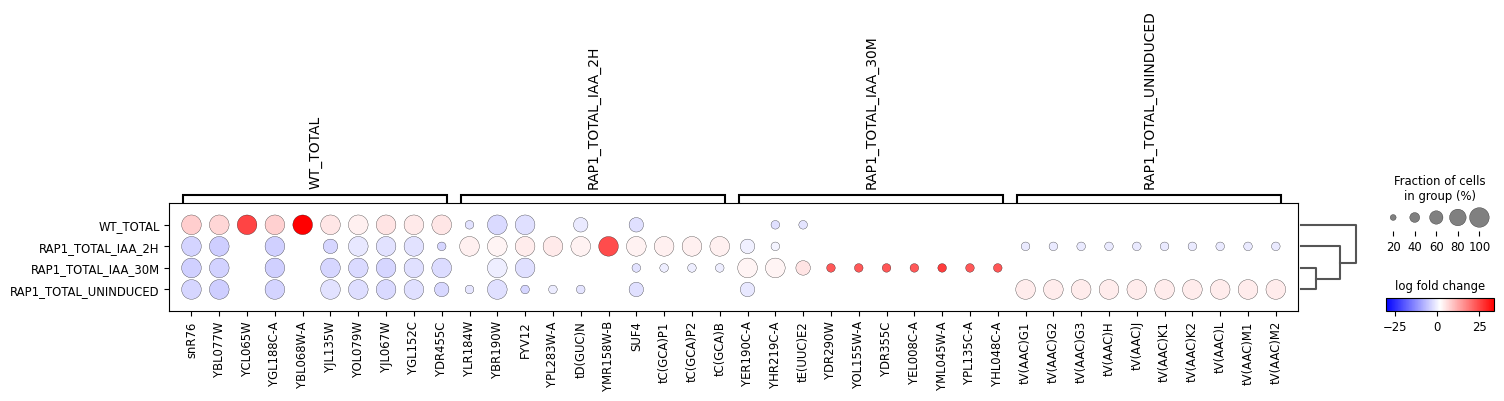
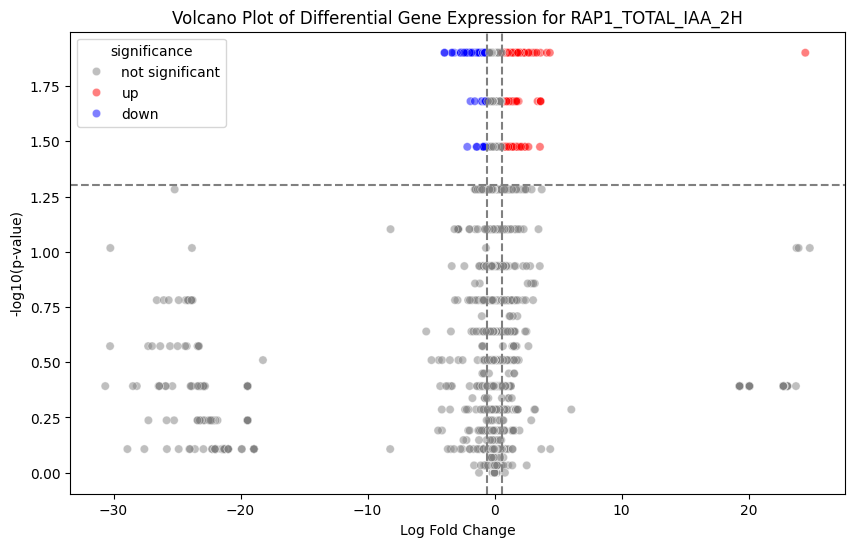
bulk-rnaseq pipeline), there a variety of tools that can be used to perform differential gene expression analysis, including the Scanpy package in Python.
Gene expression matrices, also known as count matrices, can be stored in your Mantle Database using the count_matrix data type.
In this notebook, we analyze the gene level raw counts matrix obtained using STAR and Salmon through the bulk-rnaseq pipeline. The FASTQ data were originally from:
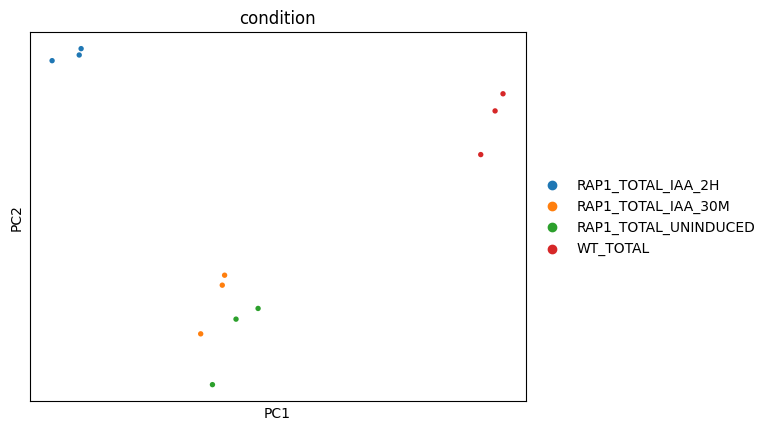
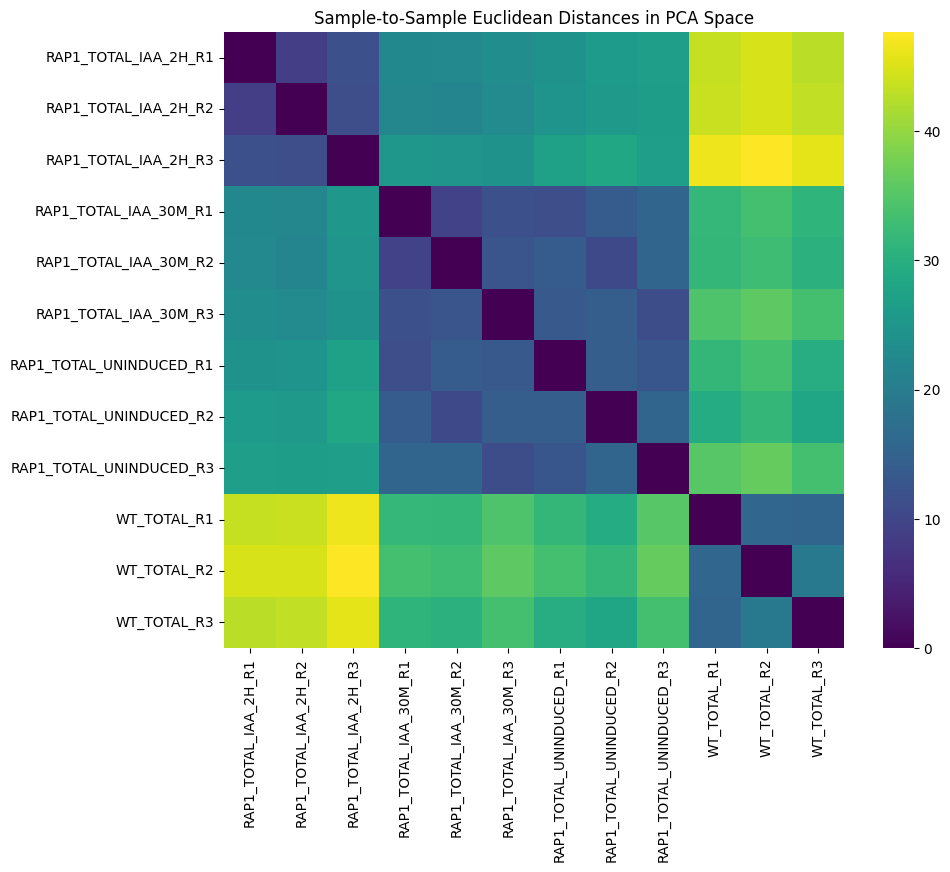
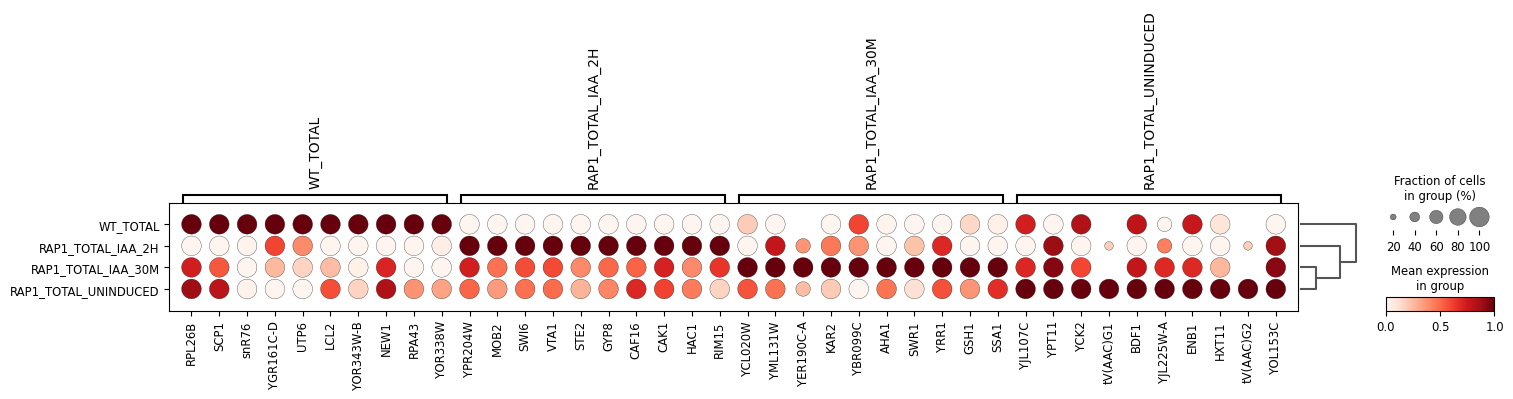
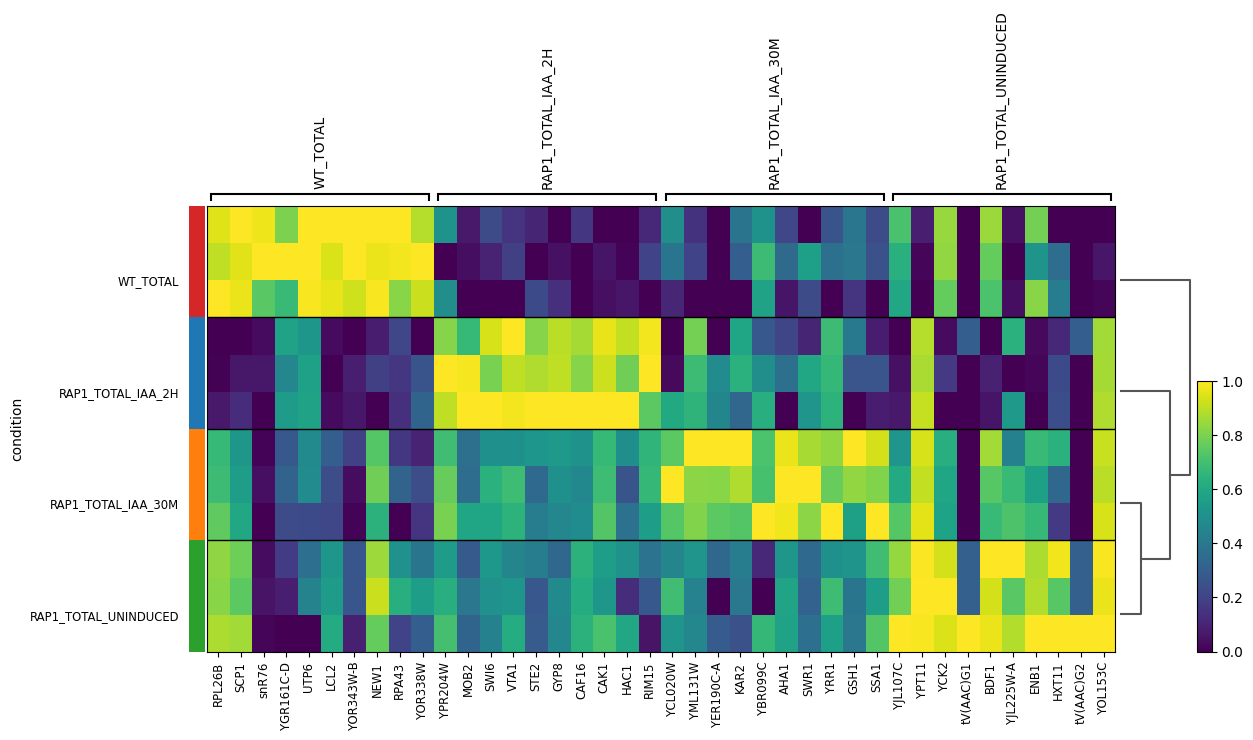
Wu, A. C. K., Patel, H., Chia, M., Moretto, F., Frith, D., Snijders, A. P., & van Werven, F. J. (2018). Repression of divergent noncoding transcription by a sequence-specific transcription factor. Molecular Cell, 72(6), 942-954.e7. https://doi.org/10.1016/j.molcel.2018.10.018Using Scanpy, we load the count matrix into AnnData format, then normalize and transform the count matrix and perform principal component analysis.