Introduction
Spatial transcriptomics is the visualization and quantification of gene expression in tissue sections, maintaining the spatial context of the tissue architecture. One technology for spatial transcriptomics is 10x Genomics Visium. It uses a slide with an array of spots, each containing barcoded oligonucleotides, to capture mRNA from tissue sections placed on the slide. The mRNA then undergoes high-throughput RNA sequencing. The raw results from Visium experiments include a microscopy image of the tissue slice on the Visium slide and sequencing reads in FASTQ format. In order to use this data to reveal insights into cellular microenvironments, cellular heterogeneity, and spatial relationships within tissue, the data require processing, including the typical steps taken for processing RNA sequencing reads, as well as tissue detection and determination of which barcode spots overlap with the tissue. 10x Genomics provides thespaceranger count pipeline for generating feature-barcode matrices, identifying clustering, and performing differential gene expression from Visium data. We have adapted spaceranger count to read and write data to your Mantle Database.
Managing Visium data and references
In your Mantle Database, data is stored as entities, which have properties that can be files or metadata. Visium experimental data can be stored in Mantle as entities of thespatial-experiment data type. Expand to view the properties of spatial-experiment entities:
Properties
Properties
Directory containing RNA sequencing FASTQ files
Microscopy image file
image, darkimage, colorizedimage, or cytaimage.Used as an input to Spaceranger Count as --<image_type> <image>.See the Spaceranger Count documentation for more information.The Visium slide serial number. Corresponds to the
slide input to Spaceranger Count. See the Spaceranger Count documentation for more information.Optional. The slide design file for your slide. See the Spaceranger Count documentation for more information.
Optional. The slide design file for your slide. Corresponds to the
loupe-alignment input to Spaceranger Count. See the Spaceranger Count documentation for more information.Visium capture area identifier. Corresponds to the
area input to Spaceranger Count. See the Spaceranger Count documentation for more information.Optional. The publication that the data originated from.
Optional. The SRA Accession Number for the FASTQ files.
Sudmeier, et al. (2022) “Distinct phenotypic states and spatial distribution of CD8+ T cell clonotypes in human brain metastases.” Cell Reports Medicine, 3(5), 100620To perform sequence alignment, a reference genome is needed. 10x has references available for human (
GRCH38-2020-A) and mouse (mm10-2020-A) genomes. They are available in your Mantle Database as spatial-reference entities.
Depending on your experimental workflow, you may also need a spatial
probe file for data processing. 10x also has probesets available for
human and mouse, which are available in your Mantle Database as spatial-probes entities.
Processing Visium data to produce feature-barcode matrices, identify clusters, and perform differential gene expression
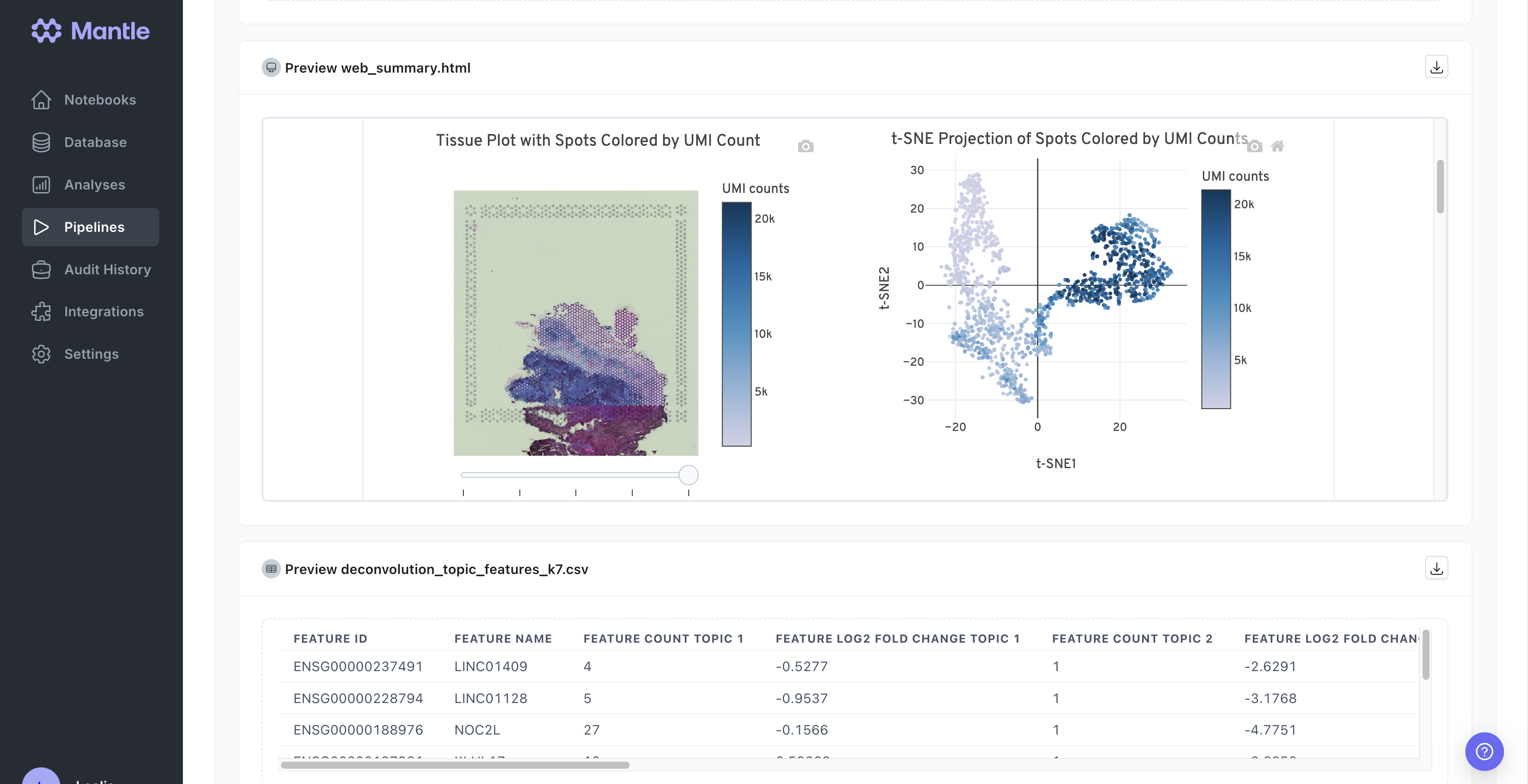
spaceranger-count pipeline adapts the 10x Genomics spaceranger count pipeline to allow it to take a Mantle spatial-experiment entity as input and to create a spaceranger-count-outputs entity with the results directory. Additionally, when you run the Mantle spaceranger-count pipeline, many of the results files can be previewed on the run page, including the an interactive pipeline report. All pipeline results files can also be browsed on the pipeline run page. Learn more about the pipeline in the 10x Genomics spaceranger count documentation.