Introduction
Mantle is a centralized solution for organizing both files and metadata, together. You can store large files (such as microscopy images or FASTQ sequencing data) alongside metadata (such as sample ID, sample origin, and experimental conditions). You can also store files together, such as the FASTQ files and image file from a spatial transcriptomics experiment. You don’t need to rely on storing all of the important descriptors in your file names.Data entities and data types
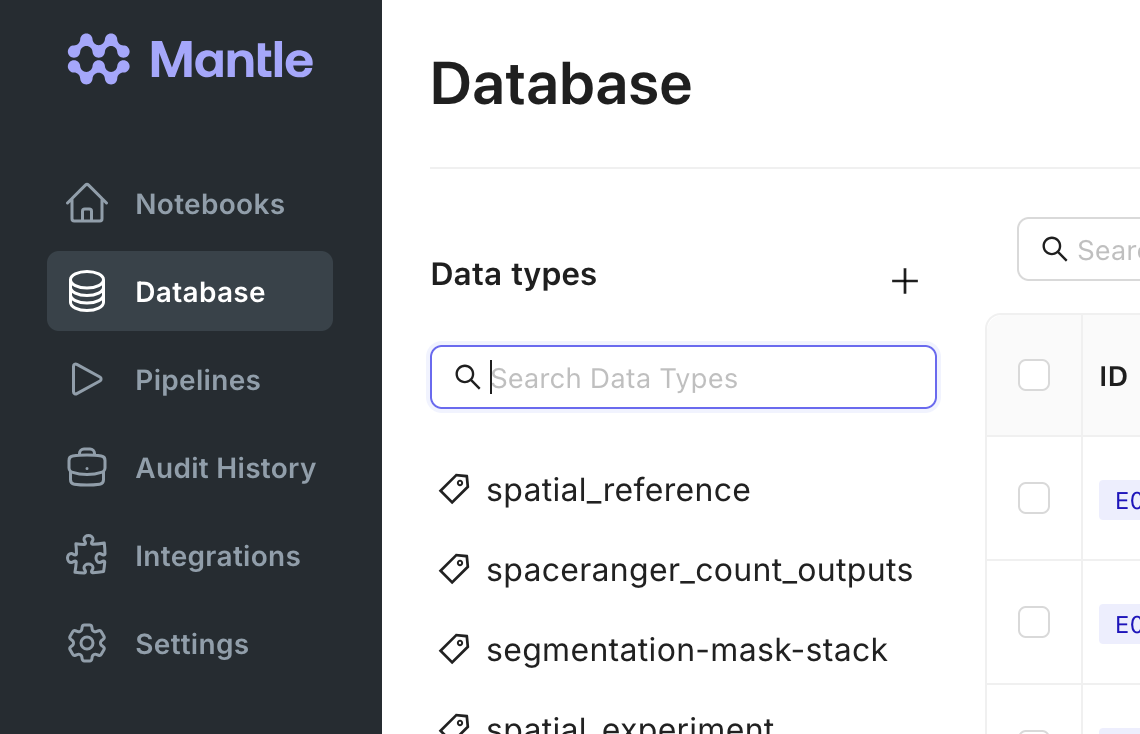
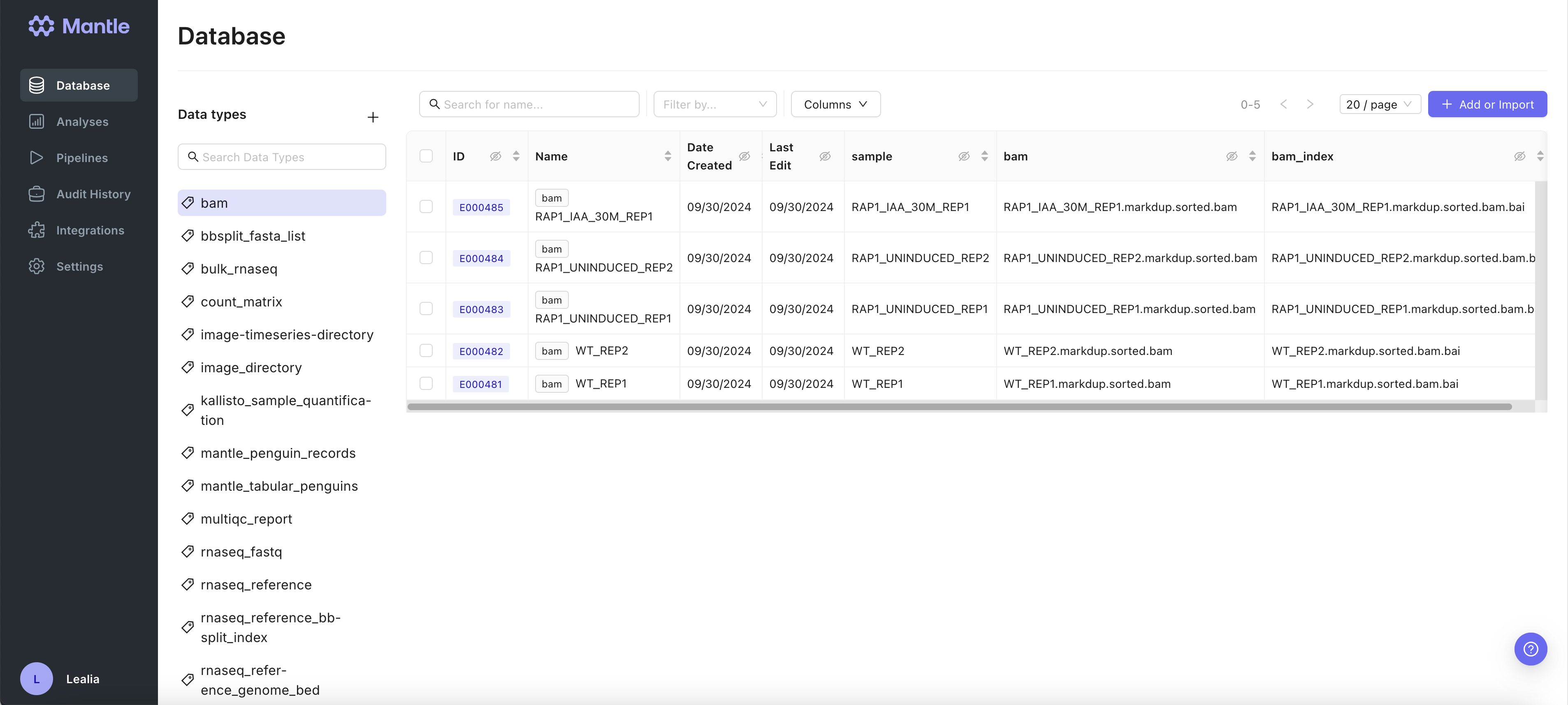
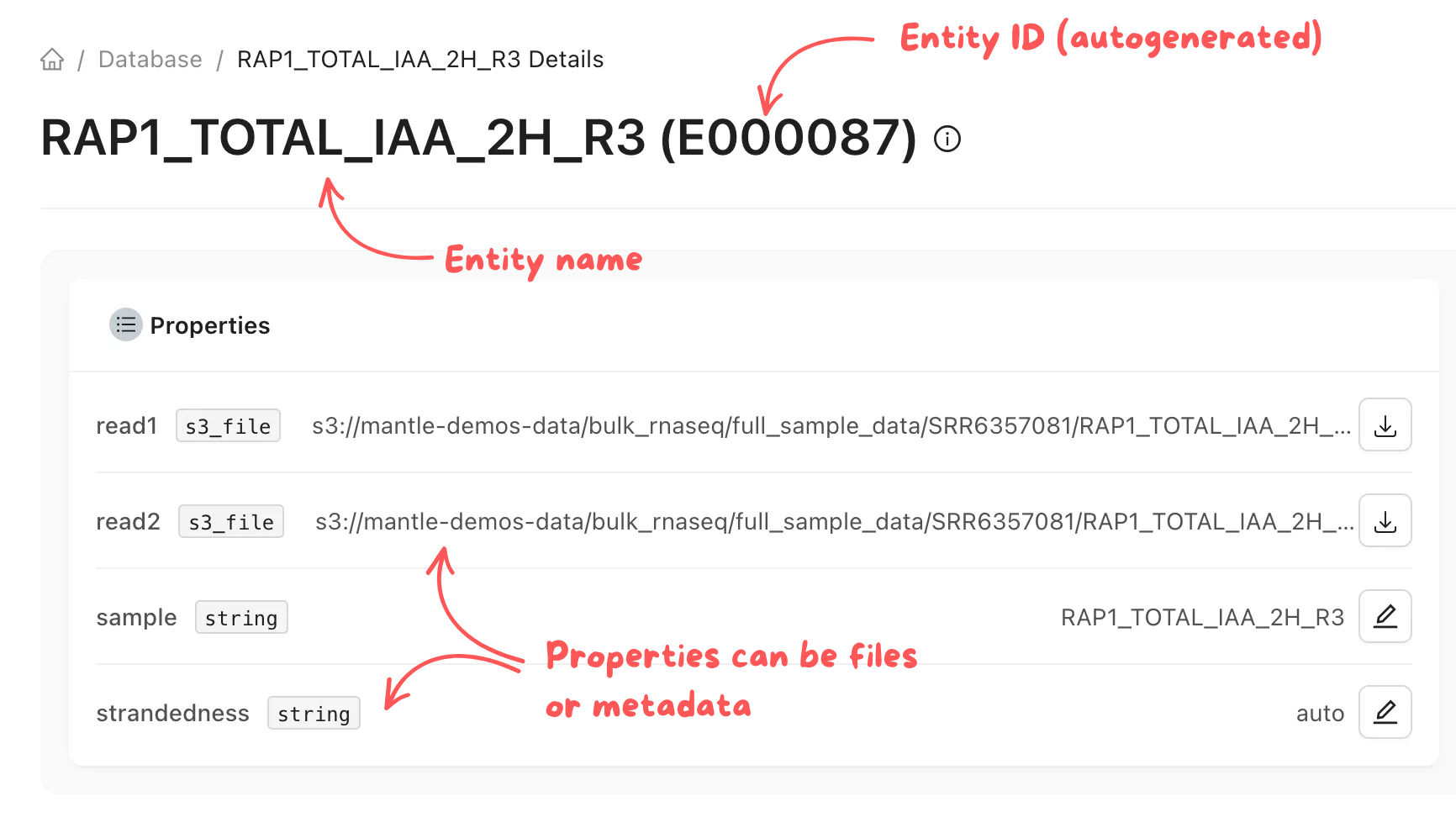
In your Mantle Database, data is stored as entities, which each have a name and one or more properties. These properties can be files or experimental metadata. Each entity is a row in a table, and the properties are columns of the table. Each table in your Mantle database is defined by a data type. There a multiple data types, such asrnaseq_fastq or image_directory.
You can browse by data type in your Mantle Database. Selecting a data type shows you all the entities of that data type in a table view.
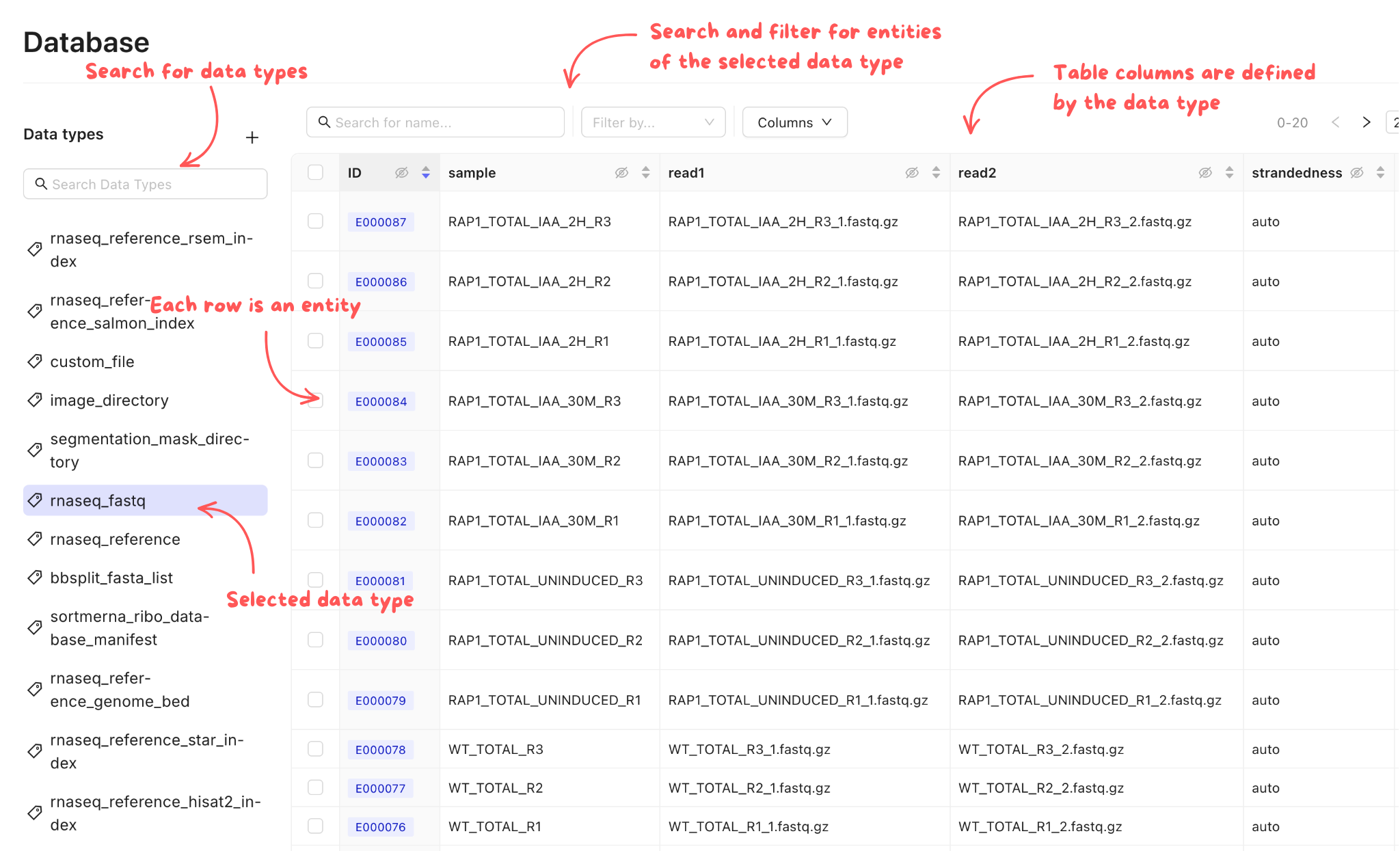
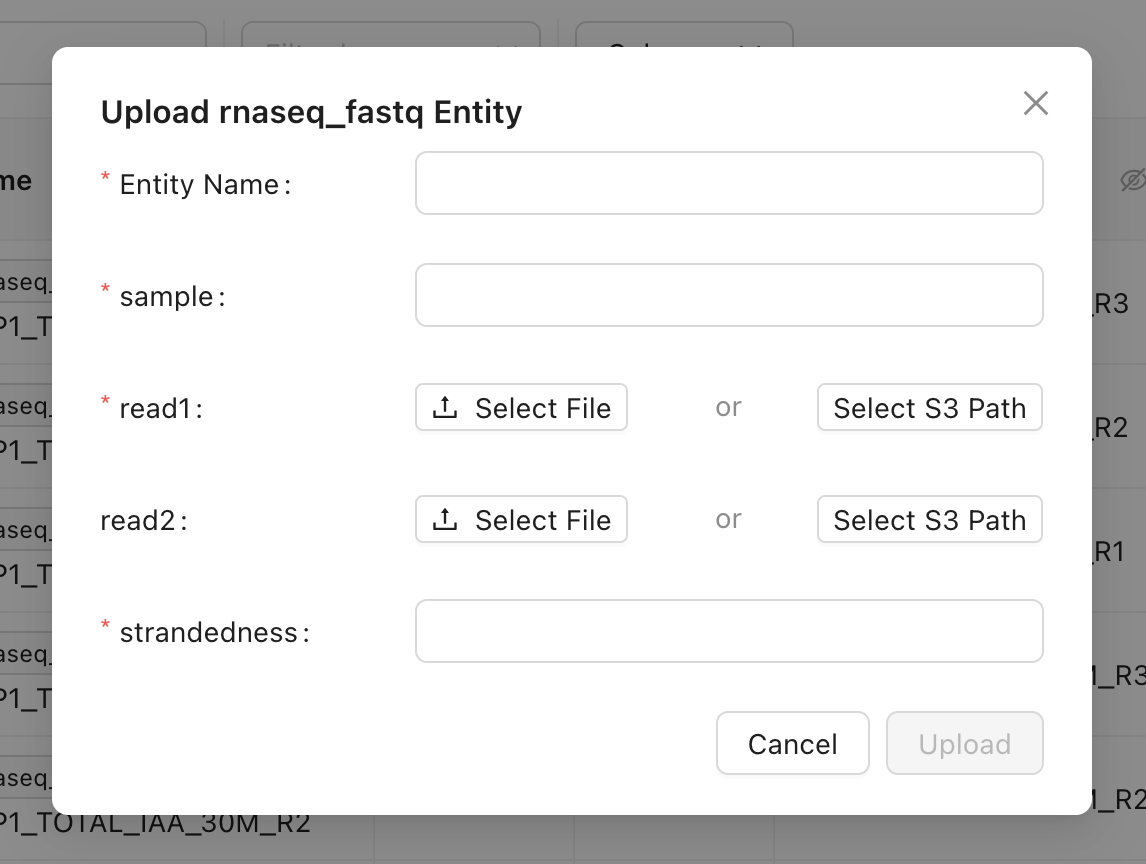
rnaseq_fastq data type contains a read1 FASTQ file, an optional read2 FASTQ file, a sample name, and a strandedness . The file properties can be downloaded to your local environment, and you can edit other properties as needed. The history of all changes is automatically tracked for you.
Adding a new data type
Your Mantle workspace comes preloaded with several data types, includingcustom_file and custom_directory types for if none of the other types apply to what you are working on. Contact us to add a new data type with custom properties.
Adding a new entity
There are three ways to add a new data entity to your Mantle Database: on the Database page, in a Mantle Notebook, or using the Mantle SDK (on or off of Mantle).On the Database page
After you refresh the page, you can view your new entity in the table.
In a Mantle Notebook
Add a Data cell to the notebook you are working in, selecting the data type for the entity you are adding.