Introduction
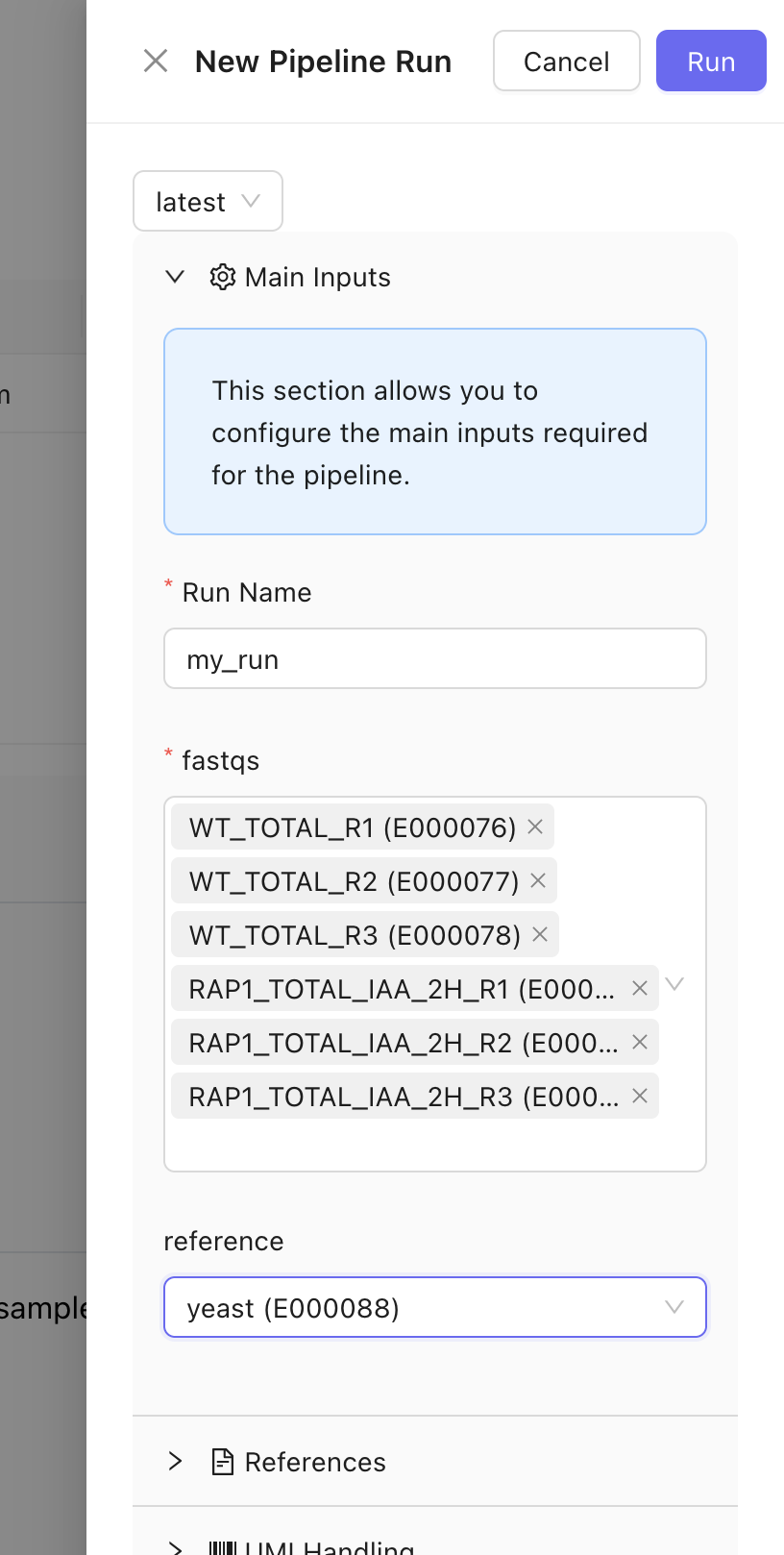
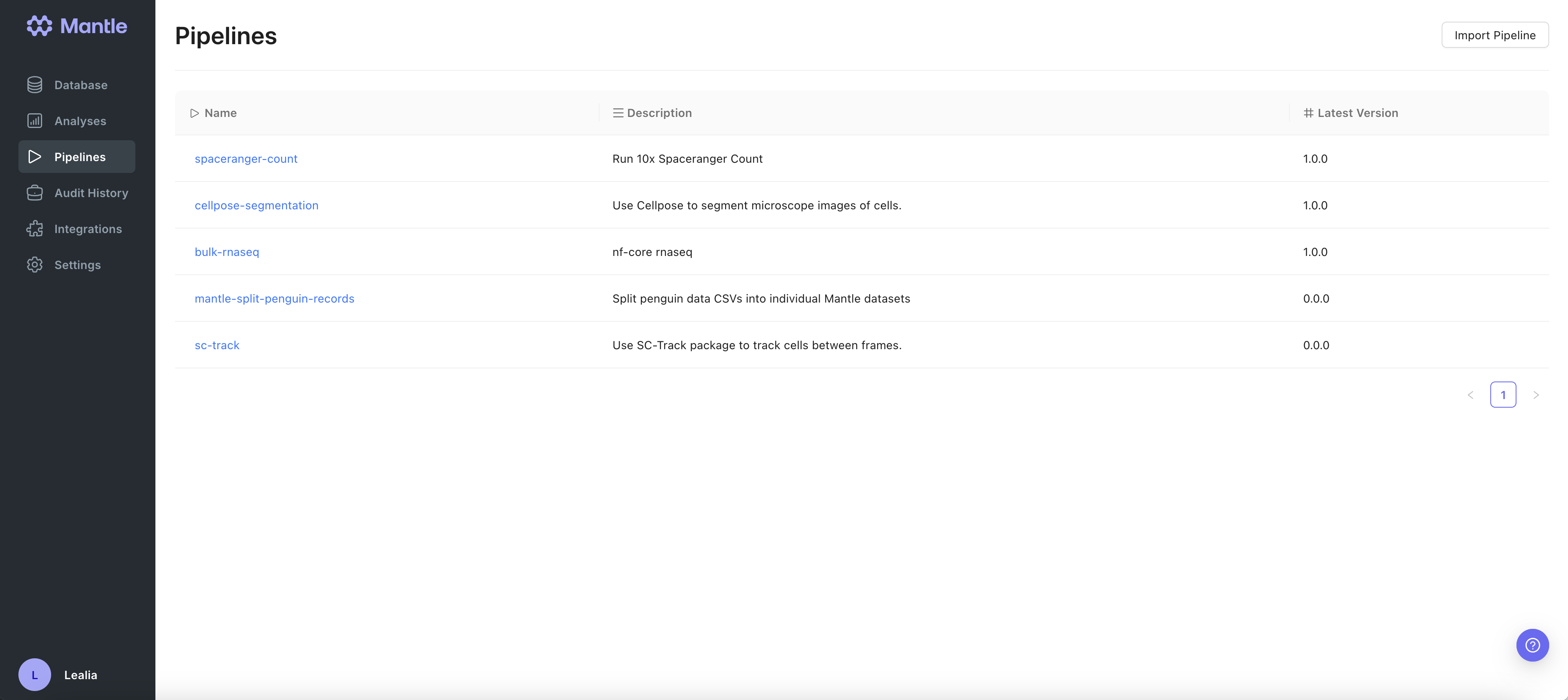
A bioinformatics pipeline is a program used to process large biological data sets, such as sequencing data. It automates data workflows, enabling reproducibility and scalability in analyses. You can run pipelines on Mantle using a user-friendly graphical interface. The pipeline runs on powerful hardware in the cloud, so you don’t have to worry about downloading anything to your computer or running out of resources on your computer. Mantle pipelines are written in Nextflow , a workflow definition system with a large open-source bioinformatics pipeline development community, nf-core.Running a pipeline
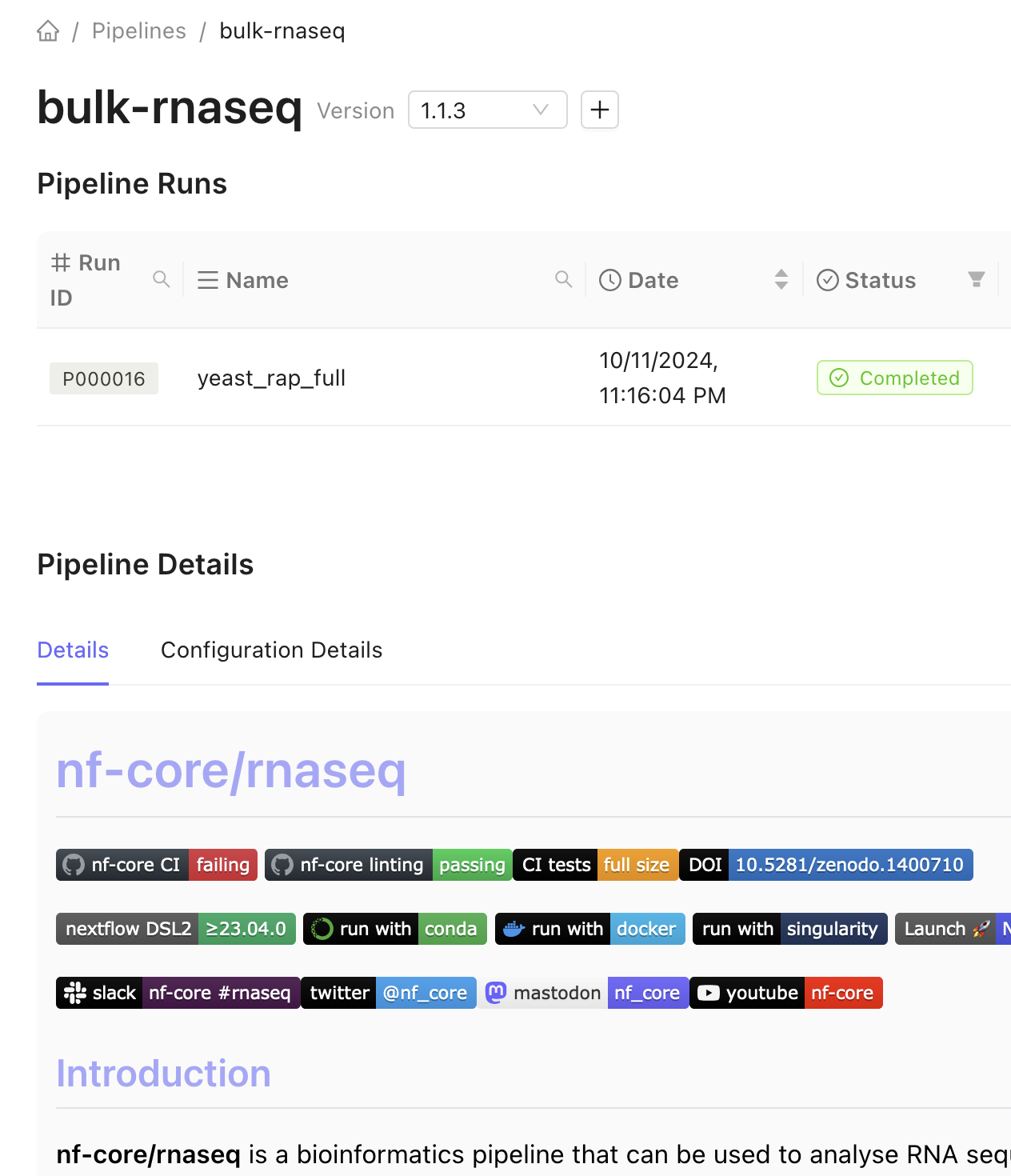
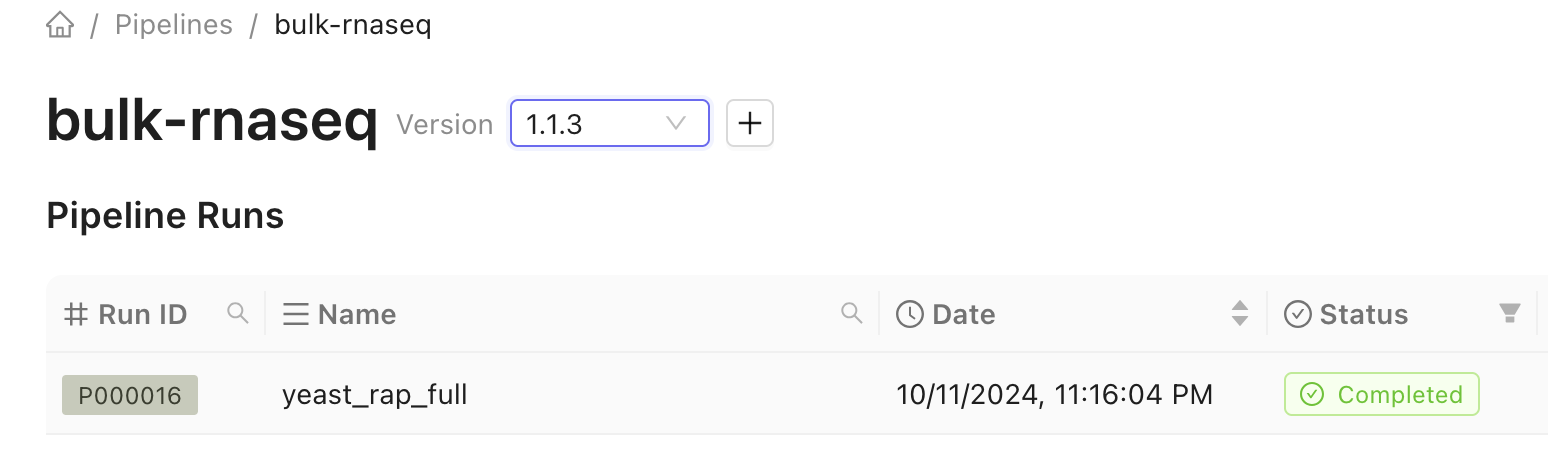
Any previous runs are displayed on in the table on the pipeline’s page.
In addition, a README file is displayed at the bottom of the pipeline’s page.